Persistent Homology Analysis of Biomolecules
Presenter
July 20, 2015
Keywords:
- Homotopy; Topological
Abstract
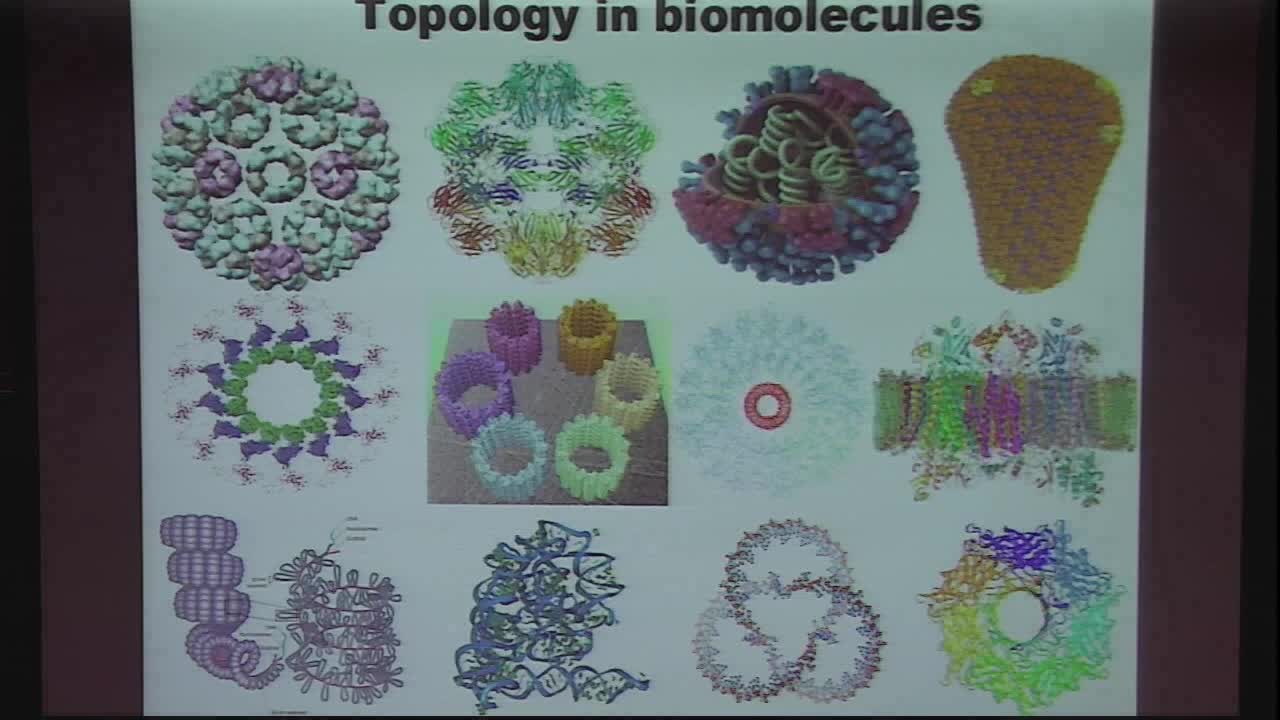
Proteins are the most important biomolecules for living
organisms. The understanding of protein structure, function, dynamics, and
transport is one of the most challenging tasks in biological science. We
have introduced persistent homology for extracting molecular topological
fingerprints (MTFs) based on the persistence of molecular topological
invariants. MTFs are utilized for protein characterization, identification,
and classification. Both all-atom and coarse-grained representations of
MTFs are constructed. On the basis of the correlation between protein
compactness, rigidity, and connectivity, we propose an accumulated bar
length generated from persistent topological invariants for the
quantitative modeling of protein flexibility. To this end, a correlation
matrix-based filtration is developed. This approach gives rise to an
accurate prediction of the optimal characteristic distance used in protein
B-factor analysis. Finally, MTFs are employed to characterize protein
topological evolution during protein folding and quantitatively predict the
protein folding stability. An excellent consistence between our persistent
homology prediction and molecular dynamics simulation is found. This work
reveals the topology-function relationship of proteins.