Brain-Scale Simulations at Cellular and Synaptic Resolution: Necessity and Feasibility - Markus Diesmann, Juelich Research Centre
August 18, 2015
Keywords:
- Interaction between neurons
- Neurons
- Cortical Layer
- Excitiatory neuron connection
- Inhibitory neuron connection
- Cortical microcircuit
Abstract
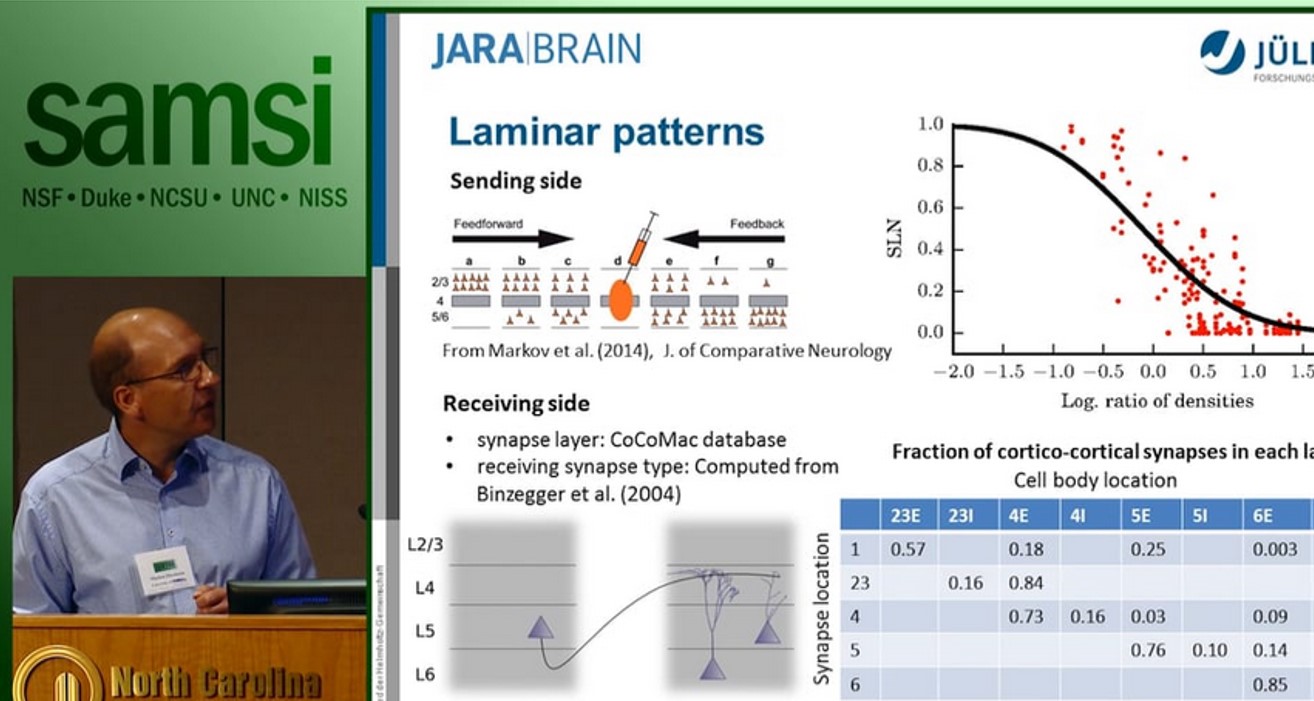
The cortical microcircuit, the network comprising a square millimeter of brain tissue, has been the subject of intense experimental and theoretical research. The lecture first introduces a full-scale model of this circuit at cellular and synaptic resolution [1]: the model comprises about 100,000 neurons and one billion local synapses connecting them. The purpose of the model is to investigate the effect of network structure on the observed activity. To this end it incorporates cell-type specific connectivity but identical single neurons dynamics for all cell types. The emerging network activity exhibits a number of the fundamental properties of in vivo activity: asynchronous irregular activity, layer specific spike rates, higher spike rates of inhibitory neurons as compared to excitatory neurons, and a characteristic response to transient input. Despite this success, the explanatory power of such local models is limited as half of the synapses of each excitatory nerve cell have non-local origins and at the level of areas the brain constitutes a recurrent network of networks.
The second part of the lecture therefore argues for the need of brain-scale models to arrive at self-consistent descriptions of the multi-scale architecture of the network. Such models will enable us to relate the microscopic activity to mesoscopic measures [2] and functional imaging data and to interpret those with respect to brain structure. Theoretical arguments support that generally networks cannot be scaled down without perturbation of their correlation structure [3].
The third part of the lecture introduces the technology required to simulate such models and discusses the performance of the present NEST simulation code. Brain-scale networks exhibit a breathtaking heterogeneity in the dynamical properties and parameters of their constituents. Over the past decade researchers have learned to manage the heterogeneity with efficient data structures [4]. Already early parallel codes had distributed target lists, consuming memory for a synapse on just one compute node. As petascale computers with some 100,000 nodes become increasingly available for neuroscience, new challenges arise: Each nerve cell contacts on the order of 10,000 other neurons and thus has targets only on a fraction of all nodes; furthermore, for any given source neuron, at most a single synapse is typically created on any node. The heterogeneity in the synaptic target lists thus collapses along two dimensions: the dimension of the types of synapses and the dimension of the number of synapses of a given type. The latest technology [5] takes advantage of this double collapse using metaprogramming techniques and orchestrates the full memory of petascale computers like JUQUEEN and the K computer into a single brain-scale simulation.