Inference of Morphogenic Pathways from Live Cell Images
Presenter
March 6, 2008
Keywords:
- Chemical
MSC:
- 74E40
Abstract
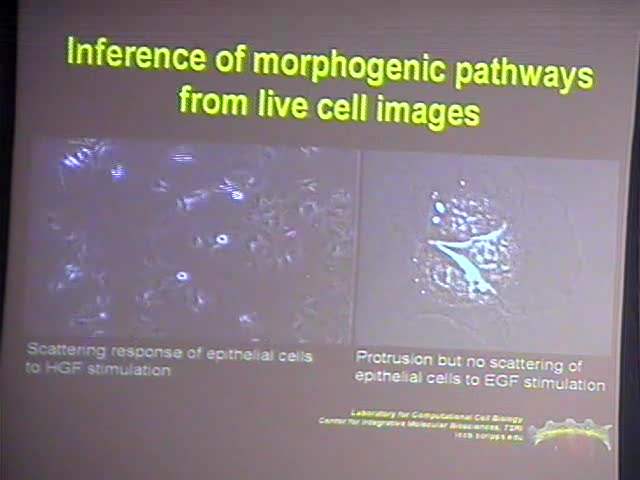
Morphogenic pathways such as those implicated in cell migration are regulated in space and time. Often they integrate mechanical signals with long-range effects and chemical signals with shorter range effects. One of the prime challenges in the analysis of pathways is to define the hierarchy and kinetics of signal transduction between spatially and temporally distributed pathway components. My lab is building a novel image analysis paradigm by which we multiplex image measurements across many experiments to register time courses of signaling events relative to cellular outputs. Indirectly, this defines also the sequence of activation of pathway components that are not simultaneously measured. Key to our multiplexing concept is the local analysis of constitutive stochastic fluctuations in pathways components with high resolution, i.e. below the diffusion radius of signaling molecules. Under these conditions, the correlation of time courses reveals precise information of the timing and of the spatial relationships between pathway activities, independent of intra-cellular and inter-cellular heterogeneity. From the timing, we can then infer causality and derive maps of the pathway hierarchy. In this presentation I will outline the idea of image fluctuation analysis and multiplexing and present first examples of pathway inference that underpin the potential of this data analysis approach.