Algebraic and topological models for DNA recombination
Presenter
November 6, 2020
Abstract
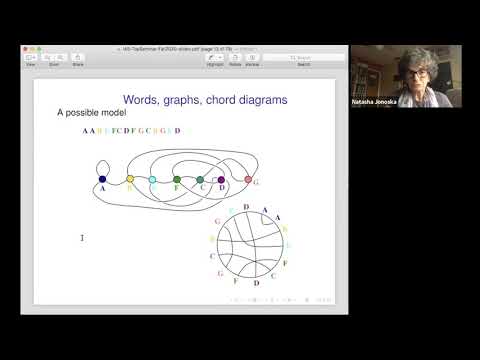
DNA rearrangement is observed at developmental and evolutionary scale. The recombination process can be directly modeled by 4-regular graphs and Gauss codes, also called double occurrence words. We discuss properties of these graphs, their spatial embedding and max genera. We illustrate these models through the recombination processes in a well studied ciliate species Oxytricha trifallax where DNA recombination is observed on a massive scale. We use word patterns within the double occurrence words to investigate genome-wide scrambled gene architectures that describe the precursor-product relationships. The genome wide rearrangement can be presented as directed graph and the first homology can be used to detect different pathways of recombination. Gene segments that recombine during these processes may be organized on the chromosome in a variety of ways. They can overlap, interleave or one may be a subsegment of another. We use colored directed graphs to represent contigs containing scrambled segment. Using graph properties to each graph we associate a point in a higher dimensional Euclidean space such that cluster formations and analysis can be performed with methods from topological data analysis. The analysis shows some emerging graph structures indicating that segments of a single gene can interleave in between, or even contain, all of the segments from several other genes.