Multiscale Representation of Macromolecular Motion
How many variables do we need to describe macromolecular motion? The answer depends on the time and length scale of interest.
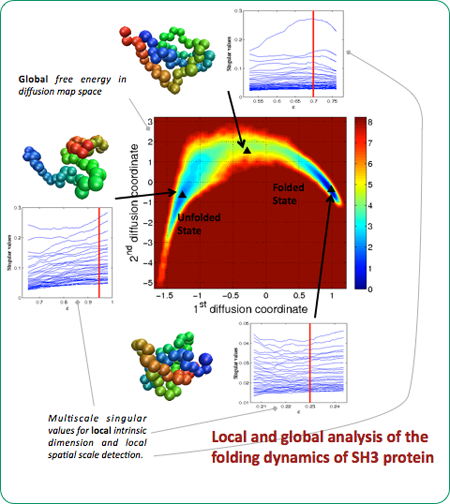
By analyzing the geometry of the space defined by long molecular dynamics trajectories of macromolecular systems, we find that different regions of the configurational space have different local intrinsic dimension and local spatial scale .
When this local heterogeneity is taken into account, it is possible to define locally scaled diffusion maps that yield global reaction coordinates for the system and associated low–dimensional nonlinear projections of the configurational space, to describe the slowest motions. This low–dimensional representation of the system on the long timescale is able to accurately reproduce certain statistics of the dynamics, such as reaction rates between semi–stable configurations. The methods proposed hold promise for larger and more complex molecules.
More details can be found in the following articles: